R Primer for Bioinformatics
R is a programming language and software that is used for statistical computation. It was developed by the R foundation in 1993 and has since become an essential tool for statisticians and data miners. Due to the large volumes and nature of the data produced in the health sciences, R is an essential skill for those in the field.
This module is meant to function as a starter kit in your journey of learning R and other programming languages.
Learning Objectives:
By the end of this module, you will
-
- Be familiar with RStudio and its functionalities
- Be able to install and use packages
- Be able to write simple scripts
- Learn about data structures and their syntax
- Learn about operators and their usage in scripts
- Be able to define and use functions
- Learn about missing values and methods to remove them
- Find patterns and perform substitutions in data using regular expressions
- Be able to subset data from a larger dataset
- Be able to read files into data frames
- Learn some basic markdown methods
- Learn about the commands required to navigate the file system
- Gain experience in plotting and modifying graphs
- Learn about Bioconductor and follow a vignette offered by them
- Gain experience in data wrangling, specifically importing raw data, the conduction of some basic statistical tests, subsetting and joining data
Installing R and RStudio on your Desktop
The best way to use R is through RStudio. Here are the installation instructions to download R and RStudio, locally.
Download and Install R:
- Go to the R-Project webpage: R-Project Link
- Click on download R
- Choose the mirror (the closest one to your current location)
- Click on the download link specific to your device (Mac OS X for Mac etc). Note: the main difference between each version is the file format. For Mac click on the .pkg file in your file system and for Windows run the .exe file
- Check your downloads and complete the installation according to the prompts
Download and Install R-Studio
- Go to the R Studio webpage: R-Studio Link
- Navigate to the bottom to find Open Source > RStudio Desktop
- Click the download link
- For MacOS, save the file to your system and drag the it to your applications folder
- For Windows run the .exe file and complete the installation instructions
RStudio is updated a couple of times a year. When a new version is available, you will be asked to update. It’s a good idea to upgrade regularly so you can take advantage of the latest and greatest features.
Datasets:
Some of the exercises in this module require additional datasets. The datasets can be found in the following google drive (Please right click and open in a new tab):
This module does not require you to download the files individually. We will be downloading them directly using some code.If you’d like to preview the files, you may choose to.
Table of Contents:
Recommended Resources:
Recommended Text: R & Bioconductor Manual By Thomas Girke @ UC Riverside. This is an excellent resource, and this module summarizes and annotates, building from their resource.
Cheatsheets:
The cheatsheets below summarize the concepts in this module in a succinct page. The information is a little dense when you look at it from a beginner’s perspective.
https://www.rstudio.com/resources/cheatsheets/
Stuck? Try these tips out!
As with any skill, you are bound to make some mistakes along the way. I find these to be the most important part of my learning experience as it prompts me to dig deeper and understand things at the fundamental level. When you come across a hurdle in R, try these tips out!
- Help from RStudio: R has a lot of built in resources to help you out. They do very similar things and choosing one of these is a matter of preference.
- Help Tab: I’ve mentioned this before, this tool is a life-saver. When I don’t really know how to use a function, I can quickly look through the manual on the help window.
- Floating ToolTip: While typing a command, R offers a quick tool tip that helps you figure out the proper syntax to use
- Help(): You can use this function to search for the manual in your console or notebook
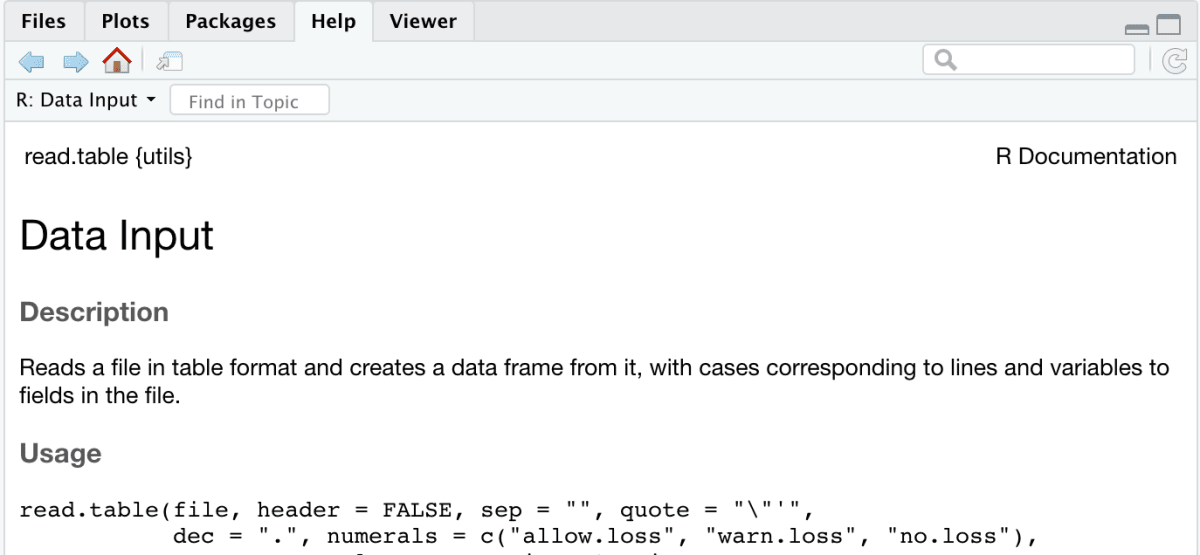
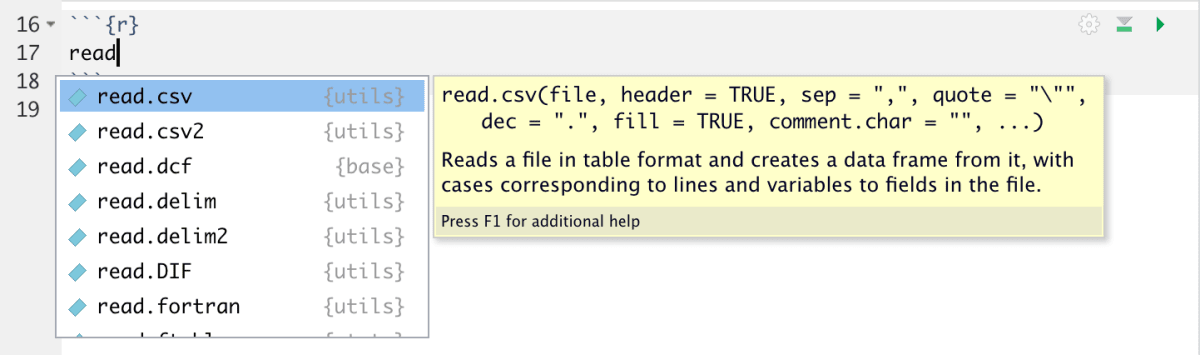
- Googling an error: When I come across an error, I copy it as it is and look for a solution on google. More often than not, someone else has had the same issue. You can see how they fixed the error and implement the same mechanism.
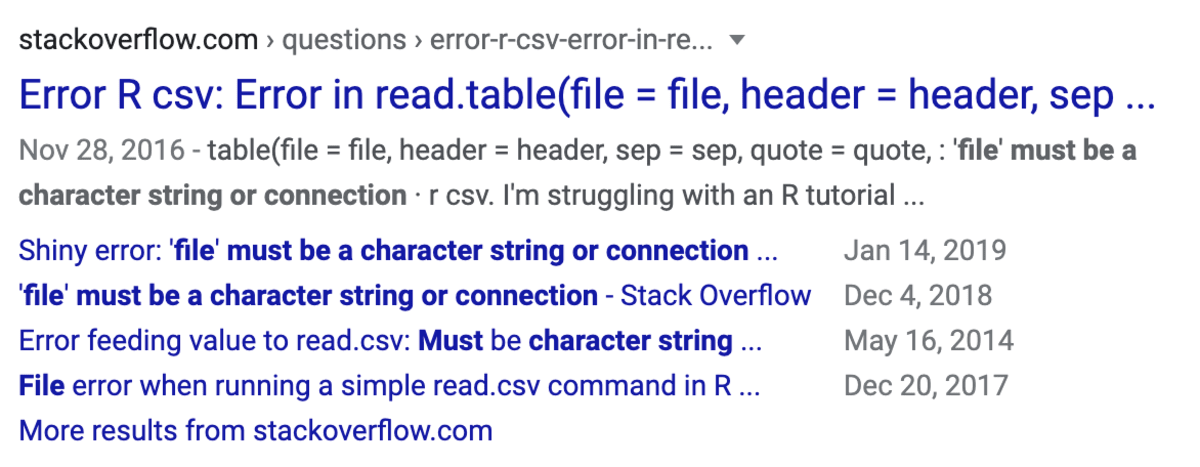
- Asking for Help- Stack Overflow: If you have a unique error that you can’t find any resources on, ask for help. You can paste your code or error message and pose a very specific question. There is a community of people who would love to help you solve the issue!